About this database
Overview
The chloroplast is the most important organelle in plants and is involved in photosynthesis and the production of numerous metabolites such as sugars, lipids, amino acids, vitamins, etc., that are important for agriculture and nutrition. However, the vast majority of the proteins in the chloroplast are encoded by the nuclear genome and are targeted to the chloroplast. Richly and Leister (2004) identified 2,090 chloroplast proteins by computational prediction of the N-terminal chloroplast transit peptides (cTPs) and demonstrated that at least 3 of the 4 different prediction softwares (iPSORT, PCLR, Predotar, and TargetP) can identify proteins with cTPs. We defined these proteins as "nuclear-encoded chloroplast proteins," and we systematically collected tagged lines that that Ds/Spm transposon or transferred DNA (T-DNA) inserted into Arabidopsis. Based on the databases of tagged mutant lines such as RIKEN Ds, JIC GT/SM, Wisconsin T-DNA, CSHL genetrap, and SALK T-DNA lines, we collected 3,246 transposon or T-DNA-tagged lines that encode 1,369 chloroplast proteins. We observed the visible phenotypes of these tagged lines on agar plates at the seedling stage; we then collected homozygous lines that had no clear phenotypes (see Methods). Mutants with heterozygotes but no homozygous knockouts were also identified in our collection, suggesting that the responsible genes of these mutants encode the essential genes in embryogenesis.
This database provides easy access to all-inclusive knowledge data, including the information of Arabidopsis knockout mutant resources and their phenotypes for nuclear-encoded chloroplast protein. Our collection of homozygous mutants, in particular, is a powerful tool for screening chloroplast mutants with cryptic abnormalities (e.g., abnormal biotic or abiotic stress response, parameters of photosynthesis, etc.) and is likely to reduce the screening time as compared to screening after ethylmethanesulfonate (EMS) mutagenesis; further, it can easily identify mutants that are yet to be identified experimentally.
If you are interested in our database or have any questions, please contact us (RARGE development team <rarge-master@psc.riken.jp>).
- RARGE: RIKEN Arabidopsis Genome Encyclopedia [Kuromori et al., 2004; Ito et al., 2005]
- EXON TRAPPING INSERT CONSORTIUM (EXOTIC) [Sundaresan et al., 1995; Tissier et al., 1999]
- WiscDsLox T-DNA Lines [Sussman et al., 2000]
- The Arabidopsis Genetrap Website at CSHL [Martienssen, 1998]
- A Sequence-Indexed Library of Insertion Mutations in the Arabidopsis Genome [Alonso et al., 2003]
- GABI-Kat: generation of flanking sequence tags (FSTs) from T-DNA mutagenised A. thaliana plants accession Columbia [Rosso et al., 2003]
- FLAGdb++: Integrative database around plant genomes [Samson et al., 2002]
Methods
The preparation of homozygous mutants and phenotype screening were performed as described below. The seeds were plated on germination medium (GM) agar plates containing 1% sucrose (Valvekens et al., 1988) with appropriate antibiotics or herbicides; they were allowed to grow in lighted growth chambers (CF-405; TOMY-Seiko, Tokyo, Japan), with approximately 75 µmol photon·mâ2·sâ1 at 22°C under a 16 h light/8 h dark cycle (long day) for 3 weeks.
With regard to the transposon-tagged lines, 60 of the seeds were harvested on the plates, and the homozygotes or heterozygotes were then selected depending on the germination rate. The strategy for producing homozygotes is illustrated in Figure 1. When the seeds obtained from heterozygote plant were grown, the segregation ratio of seedlings that are resistant or sensitive to antibiotics can be estimated at 3:1 in self crosses. On the other hand, when the seeds obtained from homozygote plant were grown, it is expected that all seedlings would be resistant to antibiotics. We examined the hereditary behavior of resistance to antibiotics in the F1âF3 (F3-F5 for RIKEN Ds) generations of the tagged lines (1 of F1, 8 of F2, and 8 of F3). In total, we examined the seeds of 16 individuals and observed their germination rates and phenotypes.
Among the T-DNA-tagged lines, the selectable marker gene cassette-encoded proteins that were resistant to antibiotics and that were integrated with the Arabidopsis genome often had multiple copies and showed complex segregation behavior with abnormal segregation ratios in their progenies. Therefore, we investigated the genotyping of individuals by using polymerase chain reaction (PCR) analysis (Figure 2). The seeds were plated on agar plates without antibiotics, and 16 randomly selected seedlings were transferred from Petri dishes to pots with soil. Several leaves were picked from the plants for DNA extraction. PCR amplification was carried out as follows: 30 s at 92°C, annealing at 55°C for 30 s, 35 cycles each at 72°C for 1 min, initial denaturation at 94°C for 1 min, and a final extension step at 72°C for 5 min. The primer pairs used for the genotyping of the F4 and F5 individuals are shown in the primer pair list file.
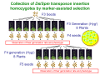 Figure 1 |
 Figure 2 |
Source information
The RIKEN Ds mutant collection described here was generated at the RIKEN Institute (Ito et al., 2002, 2005; Kuromori et al., 2004), and a part of the phenotype data of the mutants overlaps with the data in the RIKEN Arabidopsis Phenome Information Database (RAPID) (Kuromori et al., 2006). Information on other collections of insertion lines can be obtained from the T-DNA Express website. We thank all the groups and laboratories that have provided seeds from the JIC SM (Tissier et al., 1999), Wisconsin T-DNA (Sussman et al., 2000), CSHL genetrap (Martienssen, 1998), and SALK T-DNA lines (Alonso et al., 2003). To assess the linkage between mutated genes and the visible phenotype, we obtained other alleles from the SAIL T-DNA (Sessions et al., 2002), GABI-kat T-DNA (Rosso et al., 2003), and FLAG T-DNA lines (Samson et al., 2002).
- RIKEN Arabidopsis Phenome Information Database (RAPID)
- A Sequence-Indexed Library of Insertion Mutations in the Arabidopsis Genome
References cited:
- Alonso, J.M., Stepanova, A.N., Solano, R., Wisman, E., Ferrari, S., Ausubel, F.M. and Ecker, J.R. (2003) Five components of the ethylene-response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proc. Natl Acad. Sci. USA, 100, 2992-2997. [PubMed]
- Ito, T., Motohashi, R., Kuromori, T., Mizukado, S., Sakurai, T., Kanahara, H., Seki, M. and Shinozaki, K. (2002) A new resource of locally transposed Dissociation elements for screening gene-knockout lines in silico on the Arabidopsis genome. Plant Physiol. 129, 1695-1699. [PubMed]
- Ito, T., Motohashi, R., Kuromori, T., Noutoshi, Y., Seki, M., Kamiya, A., Mizukado, S., Sakurai, T. and Shinozaki, K. (2005) A resource of 5,814 dissociation transposon-tagged and sequence-indexed lines of Arabidopsis transposed from start loci on chromosome 5. Plant Cell Physiol. 46, 1149-1153. [PubMed]
- Kuromori, T., Hirayama, T., Kiyosue, Y., Takabe, H., Mizukado, S., Sakurai, T., Akiyama, K., Kamiya, A., Ito, T. and Shinozaki, K. (2004) A collection of 11 800 single-copy Ds transposon insertion lines in Arabidopsis. Plant J, 37, 897-905. [PubMed]
- Kuromori, T., Wada, T., Kamiya, A., Yuguchi, M., Yokouchi, T., Imura, Y., Takabe, H., Sakurai, T., Akiyama, K., Hirayama, T. and Shinozaki, K. (2006) A trial of phenome analysis using 4000 Ds-insertional mutants in gene-coding regions of Arabidopsis. Plant J. 47, 640-651. [PubMed]
- Martienssen R. (1998) Functional genomics: probing plant gene function and expression with transposons. Proc. Natl. Acad. Sci. 95, 2021-2026. [PubMed]
- Richly, E. and Leister, D. (2004) An improved prediction of chloroplast proteins reveals diversities and commonalities in the chloroplast proteomes of Arabidopsis and rice. Gene 329, 11-16. [PubMed]
- Rosso, M.G., Li, Y., Strizhov, N., Reiss, B., Dekker, K. and Weisshaar, B. (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol. 53, 247-259. [PubMed]
- Samson, F., Brunaud, V., Balzergue, S., Dubreucq, B., Lepiniec, L., Pelletier, G., Caboche, M. and Lecharny, A. (2002) FLAGdb/FST: a database of mapped flanking insertion sites (FSTs) of Arabidopsis thaliana T-DNA transformants. Nucleic Acids Res. 30, 94-97. [PubMed]
- Sessions, A., Burke, E., Presting, G., Aux, G., McElver, J., Patton, D., Dietrich, B., Ho, P., Bacwaden, J., Ko, C., Clarke, J.D., Cotton, D., Bullis, D., Snell, J., Miguel, T., Hutchison, D., Kimmerly, B., Mitzel, T., Katagiri, F., Glazebrook, J., Law, M. and Goff, S.A. (2002) A high-throughput Arabidopsis reverse genetics system. Plant Cell 14, 2985-94. [PubMed]
- Sundaresan, V., Springer, P., Volpe, T., Haward, S., Jones, J.D., Dean, C., Ma, H. and Martienssen, R. (1995) Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9, 1797-810. [PubMed]
- Sussman, M.R., Amasino, R.M., Young, J.C., Krysan, P.J. and Austin-Phillips, S. (2000) The Arabidopsis knockout facility at the University of Wisconsin-Madison. Plant Physiol. 124, 1465-1467. [PubMed]
- Tissier, A.F., Marillonnet, S., Klimyuk, V., Patel, K., Torres, M.A., Murphy, G. and Jones, J.D. (1999) Multiple independent defective suppressor-mutator transposon insertions in Arabidopsis: a tool for functional genomics. Plant Cell 11, 1841â1852. [PubMed]
- Valvekens, D., Van Montagu, M. and Van Lijsebettens, M. (1988) Isolation of genes expressed in specific tissues of Arabidopsis thaliana by differential screening of a genomic library. Proc. Natl. Acad. Sci. USA, 85, 5536â5540. [PubMed]


